Raster Timeseries Manager¶
The Raster Timeseries Manager is a plugin for QGIS that allows to navigate through earth observation imagery archives (e.g. Landsat, Sentinel and Modis) and derived products in space and time.

The plugin adds time controls for handling raster layers containing 4D data with two spatial dimensions, one content dimension (e.g. surface reflectance bands and derived features like indices, etc.), and one temporal dimension. Using these time controls, you can navigate and animate observations over time.
A raster timeseries is a normal raster with some additional metadata specified (see Data format).
Layer styling and spatial navigation is handled by QGIS as usual.
Table of Content
Getting started¶
- Installation
- In QGIS select ,
search for
RasterTimeseriesManagerand install the plugin. - Start
- In the toolbar click
 to show the Raster Timeseries Manager panel.
to show the Raster Timeseries Manager panel. - Load the test data
- Select Plugins > Raster Timeseries Manager > Load test data.
- Run an animation
In the Raster Timeseries Manager panel select the
timeserieslayer as current Timeseries.
Data format¶
- Overview
A timeseries composed of Earth Observation imagery is a regular 4-dimensional data cube:

With two spatial dimensions:

One spectral dimension (e.g. surface reflectance or other derived features like indices):
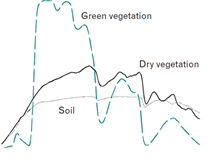
And one temporal dimension:
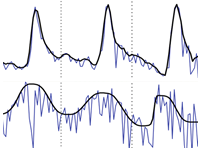
- Binary data
Data is stored as a normal raster in any GDAL supported format (e.g. GTiff, ENVI or VRT).
The raster is a stack of all bands and all observations, thus the content and temporal dimensions are flattened. This is required to match the GDAL data model, which is inherently 3d.
For example, a timeseries of ten Landsat scenes with six reflectance bands each, is stored as a raster stack with 60 bands. The first six bands belong to the first Landsat scene, the next six bands belong to the second Landsat scene, and last six bands belong to the last Landsat scene.
- Metadata
Specify names, dates and wavelength (optional) items in the TIMESERIES metadata domain.
All dates must be specified in the yyyy-MM-dd format and wavelength must be specified in Nanometers.
For the test data it looks like this:
/rastertimeseriesmanager/testdata/timeseries.bsq.aux.xml<PAMDataset> <Metadata domain="TIMESERIES"> <MDI key="names">{blue, green, red, nir, swir1, swir2}</MDI> <MDI key="dates">{1984-04-16, 1984-05-09, 1984-05-25, 1984-06-03, 1984-06-10, 1984-06-19, 1984-06-26, 1984-07-22, 1984-08-07, 1984-08-14}</MDI> <MDI key="wavelength">{483, 563, 655, 865, 1610, 2200}</MDI> </Metadata> </PAMDataset>
Tip
How to edit metadata:
- You can manually create/edit the GDAL PAM (Persitant Auxiliary Metadata) sidecar file
*.aux.xmlwith any text editor.
2. You can use the GDAL API. Note that for some raster formats, GDAL stores the metadata inside the raster file, without creating a
*.aux.xmlsidecar file:from osgeo import gdal ds = gdal.Open(filename) ds.SetMetadataItem('names', '{blue, green, red, nir, swir1, swir2}', 'TIMESERIES') ds.SetMetadataItem('dates', '{1984-04-16, 1984-05-09, 1984-05-25, 1984-06-03, 1984-06-10, 1984-06-19, 1984-06-26, 1984-07-22, 1984-08-07, 1984-08-14}', 'TIMESERIES') ds.SetMetadataItem('wavelength', '{483, 563, 655, 865, 1610, 2200}', 'TIMESERIES') ds = None
- You can manually create/edit the GDAL PAM (Persitant Auxiliary Metadata) sidecar file
Creating a timeseries¶
It can be very efficient to create a GDAL Virtual Raster File (VRT format) to specify the timeseries without duplicating the data. Virtual Raster Files can be used for raster mosaicking and stacking.
Additionally, for larger mosaics with lots of individual rasters, it is recommended to build overviews. See gdalbuildvrt and gdaladdo.
Also note the Geoserver on steroids talk about preparing large scale raster data for efficient visualisation.
Examples¶
Tasseled Cap Timeseries for whole Turkey¶
Tasseled Cap Brightness, Greenness and Wetness components for the whole area of Turkey in native 30m Landsat resolution, in 8 day steps for 2015. The dataset is presented in:
Rufin, P., Frantz, D., Ernst, S., Rabe, A., Griffiths, P., Özdoğan, M., & Hostert, P. (2019). Mapping Cropping Practices on a National Scale Using Intra-Annual Landsat Time Series Binning. Remote Sensing, 11, 232. https://doi.org/10.3390/rs11030232
Get in contact (andreas.rabe@geo.hu-berlin.de) if you want to use the dataset (~7 GB).
- Example animation zoomed in to native 30 m Landsat resolution
- Example animation zoomed to full extent
Enhanced Vegetation Index (EVI) Timeseries for the entire Brazilian savanna¶
Time series of the Enhanced Vegetation Index (EVI) for the entire Brazilian savanna
The time series covers the entire extent of the Cerrado with Landsat’s native resolution of 30 m x 30 m. The EVI values were derived from Landsat 7 and Landsat 8 observations that were acquired between April 2013 and June 2017. Data gaps in the time series were filled with an ensemble of radial basis convolution filters.
Get in contact (andreas.rabe@geo.hu-berlin.de) if you want to use the dataset (~50 GB).
- Example animation zoomed in to native 30 m Landsat resolution
- Example animation zoomed to full extent
Contact¶
Please provide feedback to Andreas Rabe (andreas.rabe@geo.hu-berlin.de) or create an issue.